Specific Process Knowledge/Lithography/EBeamLithography/BEAMER
Feedback to this page: click here
Introduction
Electron scattering in resist and substrate
During exposure electrons will undergo scattering processes in the resist and in the substrate.
Beamer
The following is a short walkthrough of the most frequently used features of Beamer. For a more in depth look at Beamer please refer to GenISys own learning material found here.
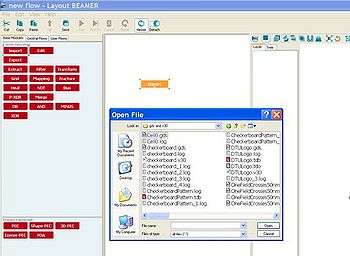
The BEAMER software is used to convert files from GDSII format to v30-format. The BEAMER software is manufactured by GenISys (www.genisys-gmbh.de) and the software is installed on the ebprep computer.
The main functions of BEAMER are:
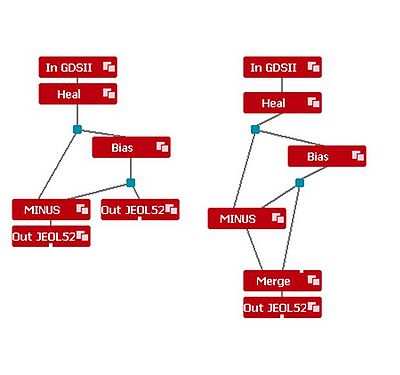
Module Function Import Imports a GDS, CIF or v30-file Export Exports to GDS, CIF or v30-format Grid Modifies the base unit of the imported file Fracture Optimizes the fracturing of the layout Heal Removes overlaps in the layout and joins abutting polygons Bias Adds or subtracts geometrical bias to the pattern Merge Merges two layouts Minus Subtracts two layouts PEC Proximity Error Correction Split Splits the output in two
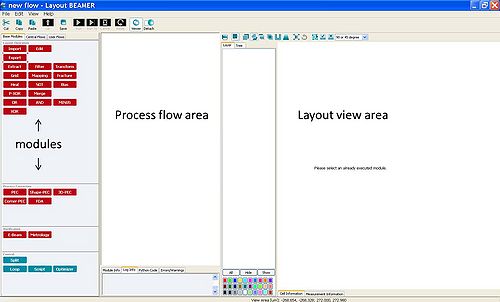
To use the modules, they are dragged to the process flow area.
The inputs and outputs of each module are illustrated by white tags. Two modules are coupled to each other by positioning the input of e.g. the Export module on top of the output of the Import module. The input/output tags turns black when they are connected.
By clicking on active modules, the layout of the file becomes visible in the layout view area.
Simple conversion from GDSII to v30
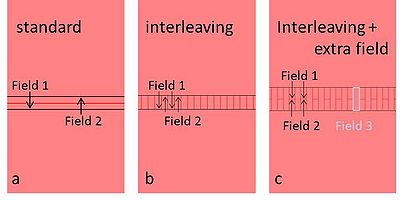
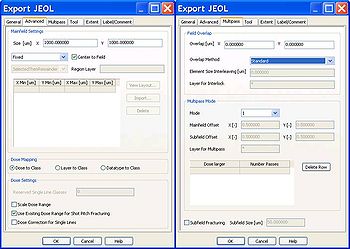
If your pattern is larger than one field size, it will be stitched by several fields. In the tab 'Multipass', you can choose to smooth the stitching errors by overlapping the fields to be stitched or by multipass method.
There are 3 types of overlap methods; for all 3 methods you define an overlap width:
a) standard: the overlap will be exposed with half dose both of field 1 and field2;
b) interleaving: the overlap will be finger-jointed by field 1 and field 2;
c) interleaving with extra field: the overlap will be finger-jointed by field 1, field 2 and an extra field 3, as illustrated to the right.
The multipass method smooths the field stitching errors by writing the pattern with half dose in different areas of the main deflector; by this method, pattern located in the corner of a writing field will be written partly by the corner of a writing field and partly by the central part of the writing field.
Dose variations
Dose variation defined by datatype or layertype
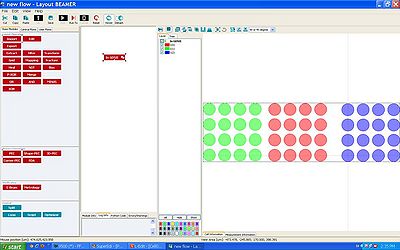
When designing your pattern, you can define a dose variation either by defining different areas in different GDS datatype or in different layertype.
In both cases you import the GDS-file as usual into BEAMER, importing all layers. The layers should be visible when you view the GDSfile:
If you defined different doses by different GDS layers, you should tick 'Datatype to Class' under 'Advanced' when you convert to v30.
If you defined different doses by different layer type, you should tick 'Layer to Class' under 'Advanced' when you convert to v30.
View the v30-file and make sure all variations have been converted correctly.
In the jdf-file, manually create a shot modulation (similar to the jdi-file created by PEC) that fits to the number of layers in your v30-file. This shot modulation should be defined in the end of the jdf-file (see the sdf- and jdf-file manual). In the example below, the three layers (green, blue and red in the BEAMER viewer to the right) are defined to have shot modulation from -20 % to + 20 % of the base-dose:
JOB/W 'TEST',4 ; 4 inch wafer
PATH DTU5M
ARRAY (0,1,0)/(10000,3,10000)
ASSIGN P(1)-> ((*,*),SHOT1)
AEND
PEND
LAYER 1
P(1) 'fredrikpnov1507prox.v30'
SPPRM 4.0,,,,1.0,1
STDCUR 0.22 ;nA
SHOT1: MODULAT (( 0, -20) , ( 1, 0) , ( 2, 20))
END
Dose variation defined by an array in jdf file
Another even faster way to define dose modulation (if you have less than 10-15 different doses) is to repeat the pattern (v30-file) in an array with different doses in the jdf file, just type the dose modulation you wish to apply to every chip in the array:
JOB/W 'TEST',4 ; 4 inch wafer
PATH DTU5M
ARRAY (-10000,2,10000)/(10000,1,10000)
ASSIGN P(1)-> ((1,1),SHOT1)
ASSIGN P(1)-> ((1,2),SHOT2)
ASSIGN P(1)-> ((2,1),SHOT3)
ASSIGN P(1)-> ((2,2),SHOT4)
AEND
PEND
LAYER 1
P(1) 'tjulahej.v30'
SPPRM 4.0,,,,1.0,1
STDCUR 0.22 ;nA
SHOT1: MODULAT (( 0, -20))
SHOT2: MODULAT (( 0, -10))
SHOT3: MODULAT (( 0, 0))
SHOT4: MODULAT (( 0, 10))
END
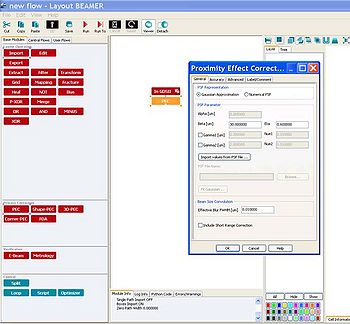
Proximity Error Correction (PEC)
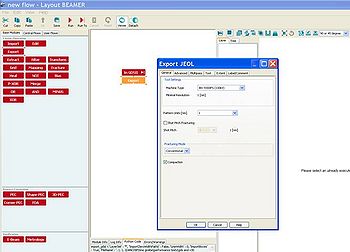
BEAMER has a built-in proximity error correction simulation program; import the GDS-file as usual and connect the 'PEC' module to its output. In the 'Proximity Error Correction' window you can either type Beta and Eta manually or import a point spread function (PSF).
Bulk and sleeve
If you have structures with both bulky and fine parts that you wish to pattern either with different doses or different currents, you can use BEAMER to divide the structures in different layers or different v30-files.
Dose test with ChipPlace
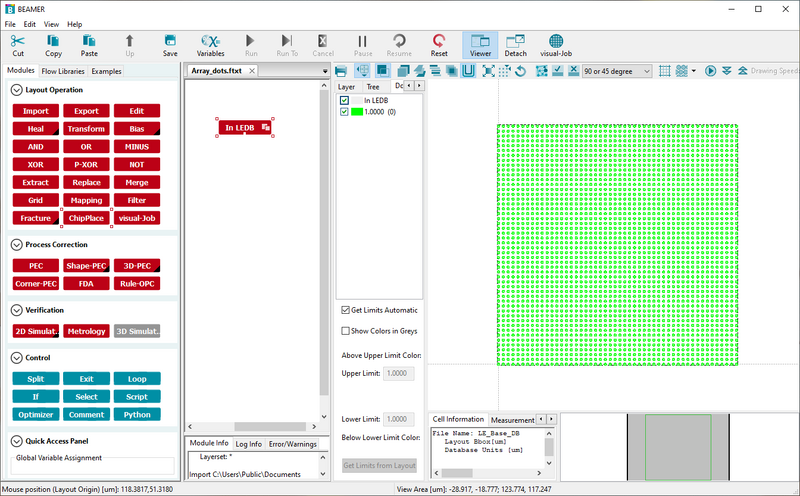
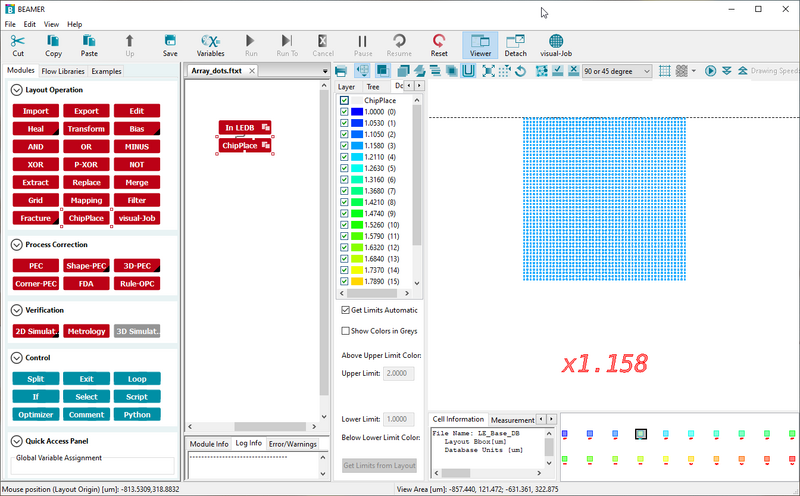
A pattern can easily be set up for a dose test using the ChipPlace module. In ChipPlace one can set up a pattern to be instanced a number of time and easily assign each instance a dose. The dose can be manually set or be linearly interpolated between a start and end value for the instanced array. The output of ChipPlace is a single pattern file with all array elements assigned to individual doses (shot ranks). This also works with patterns that already have PEC applied. In that case ChipPlace will create a combined modulation of the PEC modulation and the dose test modulation. In the example below we demonstrate how to set up dose test without PEC. If PEC is needed a PEC node can simply be added before the ChipPlace node. In the example we will use the Array_dots pattern found in the examples pane. It is a simple array of dots in a 100 x 100 µm2 box. The procedure is
- Import a design
- Add the ChipPlace node
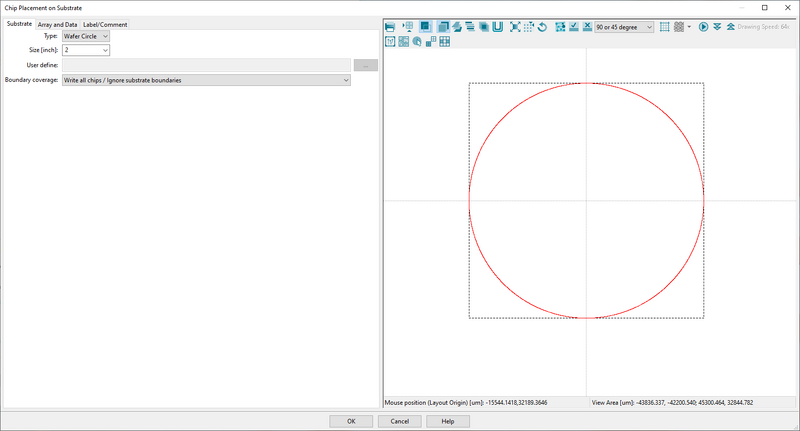
- In the Substrate tab define the substrate size. This is purely for visualization of how the pattern fits on the substrate
|
|
This example uses the Array_dots pattern supplied with Beamer. Image: Thomas Pedersen. |
|
|
Choose a substrate size to visualize placement within the substrate in ChipPlace. Image: Thomas Pedersen. |
- Go to the Array and Data tab
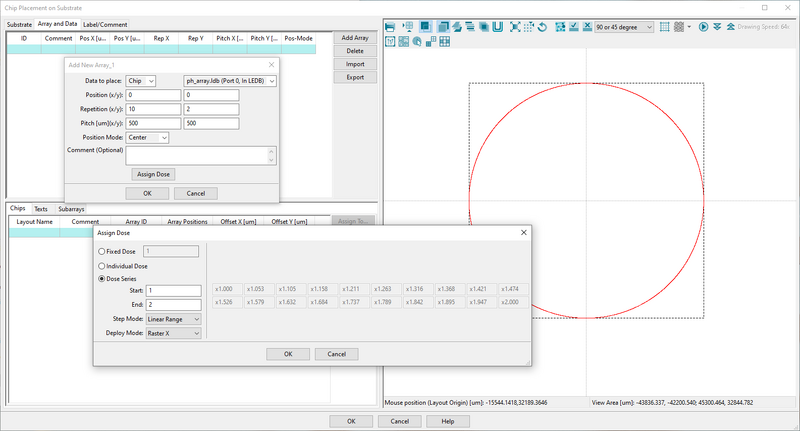
- Click Add Array
- In Data to place choose Chip. It will then take the pattern connected to the ChipPlace module
- Fill in array data in Repitition and Pitch to create as many dose test instance as you want. In this case we create a 10 x 2 array
- Choose Center in Position Mode to have everything centered in the final output pattern (V30 file)
- Click Assign Dose to open up the dose assignment page
- Choose Dose Series and enter a start and end value. This is relative dose, i.e. a multiplier on the base dose defined by RESIST in the SDF file
- Choose Raster X in Deploy Mode to increase dose from left to right for all rows
- Click OK in both open windows to see the result
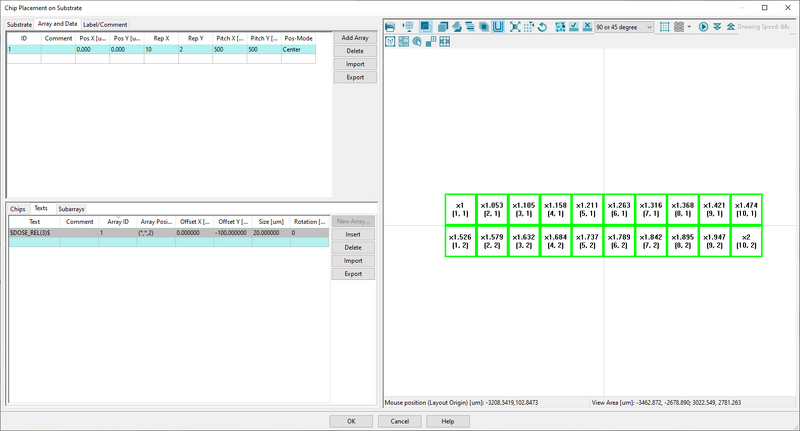
This will produce a 10 x 2 array of the pattern with a relative dose from 1 to 2. Remember this dose factor is acting as a multiplier onto the base dose defined by RESIST in the SDF file. Thus if a base dose of 200 is defined by RESIST this setup will start at 200 µC/cm2 and end at 400 µC/cm2.
| |
|
The array and dose assignment can be set up with just a few inputs. Image: Thomas Pedersen. | |
| |
|
Indicated for each element in the viewport is relative dose X and array number (x,y). Image: Thomas Pedersen. | |
To make analysis of the post exposure result easier let us add a dose label to each array element. This dose label will be exposed along with the pattern and thus when inspecting the result in SEM the (relative) dose will be visible next to the pattern. It is very easy to add:
- Go to the Texts pane in the bottom window
- Enter $DOSE_REL(3)$ to have the relative dose printed with 3 decimals accuracy
- Choose array 1 in Array ID to assign the text label to that array
- Assign text labels to all array elements with the (*,*,2) command. The "2" sets the relative dose of the labels to 2
- Assign an y-axis offset of for instance -100 µm to avoid having the label printed on top of the pattern itself
- Assign a decent size in the Size window, 20 µm will work for most cases
- Click OK in the bottom of the window to return to the Beamer view
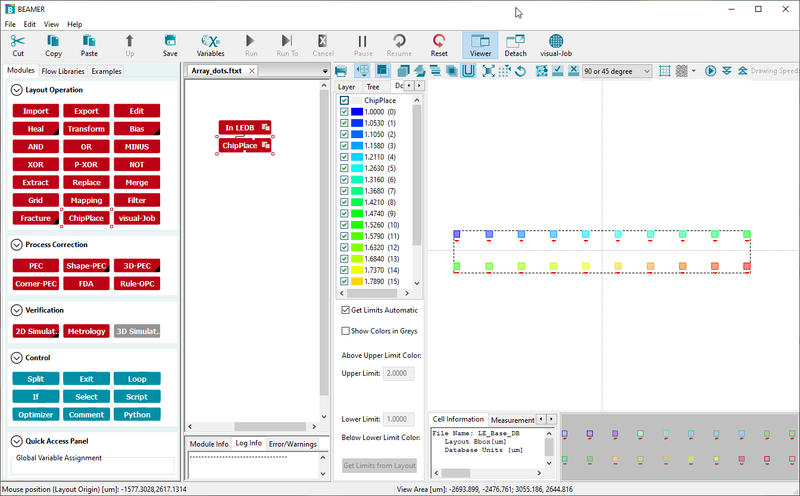
- Run the ChipPlace node to see the result
- Add an Export node and export the result as normal
|
|
Final result of the ChipPlace node with relative doses from 1 to 2, ready for export to V30. Image: Thomas Pedersen. |
|
|
Example of the added dose labels. Image: Thomas Pedersen. |