Specific Process Knowledge/Lithography/EBeamLithography/JEOLJobPreparation: Difference between revisions
| (47 intermediate revisions by one other user not shown) | |||
| Line 37: | Line 37: | ||
In the following we will discuss details of the most commonly used commands in the SDF and JDF and provide example files for various types of jobs. We will start with a simple exposure without pattern alignment, i.e. a first print exposure. In JEOL terminology this is called “mask exposure mode” whereas exposure with alignment is refered to as “direct exposure mode”. | In the following we will discuss details of the most commonly used commands in the SDF and JDF and provide example files for various types of jobs. We will start with a simple exposure without pattern alignment, i.e. a first print exposure. In JEOL terminology this is called “mask exposure mode” whereas exposure with alignment is refered to as “direct exposure mode”. | ||
Templates for SDF and JDF files can be found on the Cleanroom drive here: O:\CleanroomDrive\_JEOL9500Training\Templates | |||
=First print - mask exposure mode= | =First print - mask exposure mode= | ||
| Line 51: | Line 53: | ||
ACC 100 | ACC 100 | ||
CALPRM '2na_ap4' | CALPRM '2na_ap4' | ||
DEFMODE 2 | DEFMODE 2 | ||
FFOCUS | |||
RESIST 240 | RESIST 240 | ||
SHOT A,16 | SHOT A,16 | ||
| Line 90: | Line 93: | ||
This parameter determines if the system writes in 2 deflector mode or 1 deflector mode. In 2 deflector mode the primary deflector positions the beam within the main writing field with the subdeflector positions the beam with each 4 x 4 µm subfield. Writing speed is significantly higher in 2 deflector mode and the system should always be used in mode 2. | This parameter determines if the system writes in 2 deflector mode or 1 deflector mode. In 2 deflector mode the primary deflector positions the beam within the main writing field with the subdeflector positions the beam with each 4 x 4 µm subfield. Writing speed is significantly higher in 2 deflector mode and the system should always be used in mode 2. | ||
'''FFOCUS''' | |||
Field Focus was added to the system in 2021. It allows the system to adjust beam focus individually for each writing field based on a sample surface height map generated with HEIMAP. If FFOCUS is omitted the HEIMAP matrix will be used to calculated a single average sample height and beam focus will be set to this value. If FFOCUS is used the HEIMAP data will be used to generate a surface map and each individual writing field will be exposed with a beam focus based on this surface map. This function can not be used with height data obtained from SETWFR or CHIPAL. | |||
'''RESIST [dose]''' | '''RESIST [dose]''' | ||
| Line 187: | Line 194: | ||
Also notice that if an array element is assigned several times, it is the last assignment that will be used. | Also notice that if an array element is assigned several times, it is the last assignment that will be used. | ||
The ASSIGN command can be used to build arrays of arrays to a depth of 9 sub arrays | The ASSIGN command can be used to build arrays of arrays to a depth of 9 sub arrays. | ||
'''SKIP (j,k)''' | '''SKIP (j,k)''' | ||
| Line 217: | Line 224: | ||
The JDF is terminated with a END command. | The JDF is terminated with a END command. | ||
=Alignment exposure | =Alignment exposure= | ||
As described in the pattern preparation section there is two different strategies for pattern alignment. One can expose a wafer scale layout based on two global alignment marks or one can do chip exposures where each chip is individually aligned to an individual chip mark, as illustrated below. In both cases the SDF and JDF files have to be expanded with additional features to call and define the alignment procedure. Below is two examples, one for global alignment and one for chip alignment. Only new commands relative to the above example will be commented. | As described in the pattern preparation section there is two different strategies for pattern alignment. One can expose a wafer scale layout based on two global alignment marks or one can do chip exposures where each chip is individually aligned to an individual chip mark, as illustrated below. In both cases the SDF and JDF files have to be expanded with additional features to call and define the alignment procedure. Below is two examples, one for global alignment and one for chip alignment. Only new commands relative to the above example will be commented. | ||
| Line 225: | Line 232: | ||
|- | |- | ||
| colspan="1" style="text-align:center;| | | colspan="1" style="text-align:center;| | ||
Illustration of wafer scale pattern alignment and chip array alignment for two designs, L1 and L2. | Illustration of wafer scale pattern alignment and chip array alignment for two designs, L1 and L2. The goal is to align the L2 pattern to the L1 pattern. | ||
|} | |} | ||
==Global alignment | ==Global alignment== | ||
The following | The following files set up a single pattern for exposure with global alignment. To execute global alignment a few extra commands are used compared to the previous example. Only new commands are explained below. | ||
===Global alignment SDF=== | |||
<pre> | <pre> | ||
MAGAZIN ' | MAGAZIN 'ELELOP' | ||
#8 | #8 | ||
% 4A | % 4A | ||
JDF ' | JDF 'elelop230401',1 | ||
ACC 100 | ACC 100 | ||
CALPRM '6na_ap5' | CALPRM '6na_ap5' | ||
| Line 243: | Line 251: | ||
GLMDET A | GLMDET A | ||
CHIPAL 0 | CHIPAL 0 | ||
HSWITCH | HSWITCH ON,OFF | ||
RESIST 152 | RESIST 152 | ||
SHOT A,32 | SHOT A,32 | ||
OFFSET( | OFFSET(-150,233) | ||
END 8 | END 8 | ||
</pre> | </pre> | ||
Compared to the first print exposure the SDF file has three extra commands, '''GLMDET''', '''CHIPAL''' and '''HSWITCH'''. | Compared to the first print exposure the SDF file has three extra commands, '''GLMDET''', '''CHIPAL''' and '''HSWITCH'''. Additionally, the '''OFFSET''' command serves a somewhat different purpose now as explained below. | ||
'''GLMDET [mode]''' | |||
'''GLMDET''' (GLobal Mark DETect) has four modes | |||
*A - for automatic alignment | |||
*S - for semi automatic alignment | |||
*M - for manual alignment | |||
*C - for cancel, i.e. no alignment | |||
In automatic mode the system will execute the '''SETWFR''' subprogram to find the global alignment marks (P and Q) by beam scans. '''SETWFR''' will execute mark detection at the P and Q mark locations defined in the JDF file. If alignment fails the system will continue to the next sequence in the SDF file. In semi automatic mode the system will carry out the same alignment procedure as automatic mode but if the mark scan fails the system will ask the user to use SEM mode to manually find the mark. When the mark is found in SEM mode the user can choose to designate the position as the true mark position or request another beam scan at the found location. Thus, semi automatic is somewhat more versatile at it allows the user to manually find the mark upon failure. Manual alignment allows the user to use SEM mode and manually find and identify the location of the center of the mark. Cancel mode skips global alignment. In the example above automatic alignment is used. | |||
'''CHIPAL [mode]''' | |||
'''CHIPAL''' has six modes and is explained in the section below on chip alignment. Even though chip alignment is not used during global alignment it is necessary to include the command and set it to either 0, V1 or V4, as explained below. In the example above '''CHIPAL''' is disregarded by setting it to 0. | |||
'''HSWITCH [swg,swc]''' | |||
'''HSWITHC''' specifies the substrate height detection mode for aligned exposure. It takes two parameters '''swg''' and '''swc''' which are merely two ON/OFF switches for global mark and chip marks, respectively. If '''swg''' is set to ON the system will carry out substrate height measurement at the global mark positions. If '''swc''' is set to ON the system will carry out substrate height measurement at the chip mark positions. Thus this is a convenient way to have the system measure substrate height and adjust beam focus to chip placement positions. In the example above the global mark positions are used for height detection of the substrate. | |||
'''OFFSET[x,y]''' | |||
For aligned exposures the '''OFFSET''' command must be used to state the offset of the P mark as determined by the optical pre-aligner. This offset will be applied to the entire content of the JDF file, i.e. the offset is used to shift the mark scan detection position. In the example above the pre-aligner was used to determine the P mark is shifted -150 µm in x and 233 µm in y relative to the center of the exposure/cassette slot. | |||
==Global alignment | ===Global alignment JDF=== | ||
<pre> | <pre> | ||
JOB/W 'ELELOP',4 | JOB/W 'ELELOP',4 | ||
| Line 263: | Line 297: | ||
PATH DRF5M | PATH DRF5M | ||
ARRAY (0,1,0)/(0,1,0) ; | ARRAY (0,1,0)/(0,1,0) ; | ||
ASSIGN P(1) -> | ASSIGN P(1) -> (*,*) ; | ||
AEND | AEND | ||
PEND | PEND | ||
| Line 275: | Line 309: | ||
END | END | ||
</pre> | </pre> | ||
The JDF file has two additional commands, '''GLMPOS''' and '''GLMP'''. | |||
'''GLMPOS [P=(x1,y1), Q=(x2,y2)]''' | |||
'''GLMPPOS''' defines the global alignment mark coordinates in the substrate coordinate system and hence where '''SETWFR''' will perform beam scans for mark detection, unit is µm. In the example file the global marks are located at x = ±36000 µm and y = 0. | |||
'''GLMP [w,l,t,f]''' | |||
'''GLMP''' allows the user to add dimension information about the alignment mark and toggle between a cross and an L mark. The first parameter, '''w''', defines the width of the mark, unit is µm. The second parameter, '''l''', defines the length of the mark, unit is µm. For a cross this is the length of a single leg of the cross. The third parameter, '''t''' allows the user to toggle between a cross (0) and an L (1). The last parameter, '''f''', defines the rotation of the mark in case an L mark is used. It is most advisable to use a cross, i.e. keep the last two parameters at 0. In the example above a 2 µm line width cross with a length of 450 µm is used. | |||
==Chip alignment== | |||
Chip alignment requires a global alignment to be made first to establish the wafer coordinate system. Hence a global alignment using '''GLMDET''' is used initially as in the example above. To further illustrate chip alignment we will look at a particular layout shown below. The layout also illustrates a commonly used feature on the JEOL system to create arrays of arrays. In the layout below there is main 2x2 array and into each of these is a 5x5 subarray. Each element of the subarray is a single chip with four chip alignment marks. Notice that array placement is given in the substrate coordinate system and so is the global mark positions. Chip alignment marks (M1-M4) are however given in the local chip coordinate system. In the example files below we assume L1 is already defined on the substrate and we wish to align L2 to it. | |||
Since a pattern (V30 file) is placed at the center of the bounding box it is essential to control the bounding box of the chip design. The design in this case appears symmetric around (0,0) but in order to force it to be symmetric it is common to place corner marks at equidistance points from (0,0). The corner marks can be 1 nm boxes that will not show up in the resist when developed. | |||
{| style="border: none; border-spacing: 0; margin: 1em auto; text-align: center;" | |||
|- | |||
| [[image:EBLChipAlignExample.png|1200px]] | |||
|- | |||
| colspan="1" style="text-align:center;| | |||
Illustration of chip alignment using a chip array instanced into another array. Notice in the right most illustration how four corner marks are used to force symmetry around (0,0) such that the pattern is placed correctly. | |||
|} | |||
===Chip alignment SDF === | |||
<pre> | |||
MAGAZIN 'THOPE' | |||
#8 | |||
%4C | |||
JDF 'thope230126',1 | |||
ACC 100 | |||
CALPRM '6na_ap5' | |||
DEFMODE 2 | |||
GLMDET S | |||
CHIPAL 1 | |||
HSWITCH OFF,ON | |||
RESIST 250 | |||
SHOT A,6 | |||
OFFSET(-44,-139) | |||
END 8 | |||
</pre> | |||
'''CHIPAL [mode]''' | |||
'''CHIPAL''' has six modes | |||
*0 - Cancels chip alignment | |||
*S - SEM mode. The user is prompted to use SEM mode to manually find M1 | |||
*1 - One mark is used for position correction | |||
*4 - Four marks are used for position, rotation and gain correction | |||
*V1 - Virtual mode 1. A single mark position is used for height detection of the substrate, no position correction | |||
*V4 - Virtual mode 4. Four mark positions are used for height detection of the substrate, no position correction | |||
In addition to position correction mode 1 and 4 also detects substrate height. The virtual modes are only used to detect substrate height since no mark detection is done. Mode S obviously very time consuming for a high number of chips. | |||
If set up properly on good quality marks mode 1 or mode 4 chip alignment can usually execute in about 1-2 seconds per mark. The time estimate at compilation will account for the time spend on chip alignment at the current settings of the '''CHIPAL''' subprogram. | |||
===Chip alignment JDF === | |||
In addition to the use of '''CHMPOS''' for chip mark position definition the example below illustrate making arrays of an array. The first array is set up as a 2x2 array assigning array '''A1'''. '''A1''' is defined below as '''1:''', since '''1''' is defined as an array it can be referenced as '''A1'''. '''A''' then defines a 5x5 array assigning pattern '''P(1)''' to each element. The chip mark position command must be used in the same array that assigns the corresponding pattern. | |||
<pre> | |||
JOB/W 'THOPE',4 ; 4inch wafer | |||
GLMPOS P=(-30000,0),Q=(30000,0) | |||
PATH FT01 | |||
ARRAY (-15000,2,30000)/(10000,2,20000) | |||
ASSIGN A(1) -> ((1,1)) | |||
1: ARRAY (-4000,5,2000)/(4000,5,2000) | |||
CHMPOS M1=(-450,450),M2=(450,450),M3=(450,-450),M4=(-450,-450) | |||
ASSIGN P(1) -> (*,*) | |||
AEND | |||
PEND | |||
LAYER 1 | |||
P(1) 'thope230126.v30' | |||
SPPRM 4.0,,,,1.0,1 | |||
STDCUR 6 | |||
END | |||
</pre> | |||
'''CHMPOS [M1=(x1,y1){,M2=(x2,y2),M3=(x3,y3),M4=(x4,y4)}]''' | |||
'''CHMPOS''' defines the chip alignment marks in the local chip coordinate system, unit is µm. M1 is mandatory while M2-M4 are optional in one mark mode. In four mark mode all marks must be defined. The order of the marks is important, M1 must be top left with M2-M4 placed clockwise around the center. In the given example the four marks are placed symmetrically at ±450 µm in x and ±450 µm in y. | |||
It V1 mode it is customary to set M1 = (0,0) such that substrate height is detected at the center of the chip. In this way V1 mode can be used to exactly read out substrate height where the chip pattern will be written. | |||
=Beam current and condition files= | |||
The beam current can in principle be changed in very fine steps, it however requires recalibration of the Dynamic Focus and Dynamic Stigmation table. Hence, only a limited number of beam currents are available. The available beam currents and condition file name are listed below. | |||
{| class="wikitable" | |||
|+ Condition files and beam current | |||
|- | |||
! Beam current [nA] !! Aperture !! Condition file | |||
|- | |||
| 0.12 || 4 || 0.12na_ap4 | |||
|- | |||
| 0.16 || 4 || 0.16na_ap4 | |||
|- | |||
| 0.22 || 4 || 0.22na_ap4 | |||
|- | |||
| 0.4 || 4 || 0.4na_ap4 | |||
|- | |||
| 0.5 || 4 || 0.5na_ap4 | |||
|- | |||
| 0.8 || 4 || 0.8na_ap4 | |||
|- | |||
| 1.4 || 4 || 1.4na_ap4 | |||
|- | |||
| 1.6 || 4 || 1.6na_ap4 | |||
|- | |||
| 2 || 4 || 2na_ap4 | |||
|- | |||
| 2.7 || 4 || 2.7na_ap4 | |||
|- | |||
| 3.8 || 5 || 3.8na_ap5 | |||
|- | |||
| 4 || 4 || 4na_ap4 | |||
|- | |||
| 5 || 5 || 5na_ap5 | |||
|- | |||
| 6 || 5 || 6na_ap5 | |||
|- | |||
| 10 || 6 || 10na_ap6 | |||
|- | |||
| 12 || 5 || 12na_ap5 | |||
|- | |||
| 14 || 8 || 14na_ap8 | |||
|- | |||
| 19 || 7 || 19na_ap7 | |||
|- | |||
| 21 || 7 || 21na_ap7 | |||
|- | |||
| 22 || 7 || 22na_ap7 | |||
|- | |||
| 25 || 7 || 25na_ap7 | |||
|- | |||
| 27 || 7 || 27na_ap7 | |||
|- | |||
| 29 || 7 || 29na_ap7 | |||
|- | |||
| 30 || 8 || 30na_ap8 | |||
|- | |||
| 36 || 8 || 36na_ap8 | |||
|- | |||
| 41 || 8 || 41na_ap8 | |||
|- | |||
| 44 || 8 || 44na_ap8 | |||
|- | |||
| 54 || 7 || 54na_ap7 | |||
|- | |||
| 60 || 8 || 60na_ap8 | |||
|} | |||
Latest revision as of 09:07, 3 June 2025
Introduction
In order to write a pattern with the e-beam system you will have to generate a set of job files. These files are essentially a set of instructions which tell the system what pattern to write, where to place each pattern along with specific writing conditions, exposure dose, alignment conditions and calibration conditions. Apart from this you will naturally also have to generate the pattern you wish to expose.
The JEOL system uses the proprietary EXPRESS (EXposure PREparation System for Scheduling) system for job (schedule) definition. This system uses a compiled architecture in which all files must be prepared in advance and in turn be compiled into an executable job that can then be executed during an exposure session. From a user perspective this means that there is a significant amount of work to be done up front before any exposure can take place. It also means that the learning curve is fairly steep as users have to learn a significant amount about job file preparation in order to carry out even simple exposures. The benefit is however that at runtime, execution is relatively simple with a high likelihood of success, provided that all the preparatory work has been done correctly. At DTU Nanolab it is a strict requirement that all job files are prepared ahead of an exposure session. The JEOL 9500 system is a valuable resource and users must always show up well prepared not to waste tool time on file preparation. Users who squander valuable tool time away due to poor preparation will lose access to the tool.
JEOL 9500 file types and generalized flow
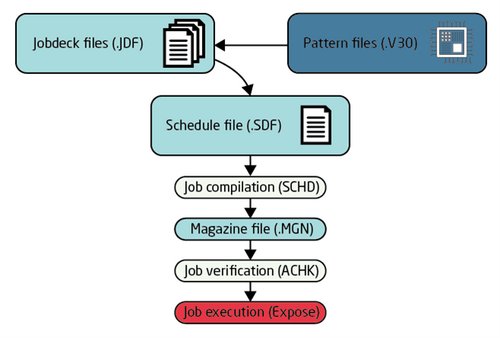
In order to execute pattern writing the job must be defined in a Schedule file (SDF) and at least one Jobdeck file (JDF). Both are text files. A pattern file defining the layout is naturally also necessary.
The SDF file contains high level job information and references a Jobdeck file (JDF) for further job information. The SDF file should contain the following information:
- Substrate cassette to use for the exposure
- Beam current and calibration parameter file to use
- Alignment type, if applicable
- Exposure dose
- Beam pitch
- Reference to Jobdeck file (JDF) with pattern placement information
The SDF will reference one or more JDF and each of these should contain the following information:
- Substrate slot ID to expose
- Initial and cyclic calibration path
- Pattern array definition, i.e. number of pattern instances and position
- Alignment mark information if alignment is used
- Reference to pattern file(s) (.V30)
- Dose modulation table if applicable
In this way the SDF is the governing file that calls a number of JDF that in turn will call and specify a pattern or sets of patterns. In order to execute the job the files must be compiled into a magazine file, this is done using the SCHD command in the command prompt on the EBL computer. After compilation the resulting magazine file should be visually validated to some degree to ensure the pattern and placement appears to be correct, this is done with the ACHK program of the system. Finally the job can be executed to expose the substrate(s).
Bear in mind that this is a generalized job flow intended to illustrate the flow with a minimum of complexity. There are several exceptions to the above and several job parameters are interchangeable between SDF and JDF.
The system is extremely syntax intolerant. The smallest syntax error will result in a compilation error and no magazine file will be generated. The error messages provided by the compiler can be hard to understand and thus it can sometimes be time consuming and frustrating to compile a working magazine file. This is another reason while a working set of job files should always be generated ahead of an actual exposure session. Remember, an exposure session is simply execution of a well prepared job.
In the following we will discuss details of the most commonly used commands in the SDF and JDF and provide example files for various types of jobs. We will start with a simple exposure without pattern alignment, i.e. a first print exposure. In JEOL terminology this is called “mask exposure mode” whereas exposure with alignment is refered to as “direct exposure mode”.
Templates for SDF and JDF files can be found on the Cleanroom drive here: O:\CleanroomDrive\_JEOL9500Training\Templates
First print - mask exposure mode
Schedule File
A simple example of a SDF is shown below. It has a single sequence and hence it will expose a single substrate. A detailed description of the commands is given below. Comments can be added using semicolon, as illustrated in the first line.
;thope220101.sdf example file
MAGAZIN 'MYWAFER'
#8
%4A
JDF 'thope220101',1
ACC 100
CALPRM '2na_ap4'
DEFMODE 2
FFOCUS
RESIST 240
SHOT A,16
OFFSET(-5000,1000)
END 8
The SDF has a three sections; a header section, a sequence section and a footer section. The job information is found in the sequence sections and there can be multiple sequences defined. Usually one sequence will be used per substrate or per beam current for multi current jobs.
SDF header section
MAGAZIN ['name’]
The SDF starts with specifying the name of the job. The name must be uppercase alphanumeric and limited to 9 characters. First character must be a letter. The name will appear in the exposure job list upon execution, but it is hardly important what it is. We recommend using your initials. In the example above the job will be named “MYWAFER”.
SDF sequence section
#[cassette number]
Specification of the cassette number to expose, i.e. from which slot of the autoloader the cassette is loaded from. This must naturally be the cassette your substrate is loaded into. In the above example the exposure will be carried out on the cassette from autoloader slot number 8.
%[slot ID]
Some cassettes have multiple substrate slots. This designates which slot of the cassette to expose. 6” and 8" cassettes only have a single slot but one must still designate that slot. In the example slot 4A will be exposed. This is slot A of a 4” wafer cassette.
JDF [‘name’],[layer number]
This specifies the name of the JDF for the exposure and the layer number of the JDF to use. The JDF can be set up to have multiple layers typically used to specify different beam currents to various designs. Please note this “layer” has nothing to do with layers of the pattern file. We advise users to name their JDF with user initials followed by exposure date. In the example the JDF called is “thope220101.jdf” using layer 1.
ACC [acceleration voltage]
This designates the acceleration voltage to be used, unit is kV. While the system can in principle run at other acceleration voltages the stabilization time is prohibitively long and thus the system is only used at 100 kV. This must always be stated as ACC 100 as in the example.
CALPRM [‘calibration file name’]
The calibration parameter command designates which calibration file to use for the exposure. This sets the exposure current and aperture used. In the example exposure will be done at 2 nA using aperture 4. It is important to stress that one cannot simply choose another beam current by writing another number here. The command references a pre-defined calibration file on the system and only a selected set of calibration files exist. These can be found and listed in the CALIB window on the system.
DEFMODE [number]
This parameter determines if the system writes in 2 deflector mode or 1 deflector mode. In 2 deflector mode the primary deflector positions the beam within the main writing field with the subdeflector positions the beam with each 4 x 4 µm subfield. Writing speed is significantly higher in 2 deflector mode and the system should always be used in mode 2.
FFOCUS
Field Focus was added to the system in 2021. It allows the system to adjust beam focus individually for each writing field based on a sample surface height map generated with HEIMAP. If FFOCUS is omitted the HEIMAP matrix will be used to calculated a single average sample height and beam focus will be set to this value. If FFOCUS is used the HEIMAP data will be used to generate a surface map and each individual writing field will be exposed with a beam focus based on this surface map. This function can not be used with height data obtained from SETWFR or CHIPAL.
RESIST [dose]
This determines the exposure dose in units of µC/cm2. The beam dwell time is automatically adjusted to achieve this area dose based on the latest beam current measurement. In the example a dose of 240 µC/cm2 is used. In many cases it can be necessary to perform a dose test to get the optimum result. A dose test can be set up in several ways, please consult with the dose testing section in Labadviser.
SHOT A,[number]
The SHOT A command determines the beam pitch, i.e. the distance between beam positions when filling in a pattern. The beam pitch grid is 0.25 nm and hence the number stated here signifies the beam pitch in units of 0.25 nm. In the example a beam pitch of 16 units will result in a beam pitch of 4 nm.
OFFSET[x,y]
The OFFSET command can be used to shift the position of the entire content of the referenced JDF, unit is µm. In the example the pattern specified in the JDF is shifted 5000 µm left and 1000 µm up. For first print (unaligned) exposures one can use the offset command to move patterns as wanted. For exposure with alignment this is however not the case, for aligned exposures the offset command must be used to state the offset of the pattern from the substrate center as determined by the optical pre-aligner.
END [number]
The END command serves two purposes. First of all it tells the software that this is the end of the job. Secondly, the number after the END command determines which cassette to load from the autoloader to the stage after job execution. In the example a pattern is written on cassette 8 and since the END command also ends with 8, no cassette change is made. If it had terminated with END 1, cassette 8 would be transferred out and cassette 1 would be transferred in after pattern writing. If the job terminates with an END command without a number, the current cassette will be unloaded and the stage left empty. This is not allowed since the stage will loose position tracking after extended time without a cassette on stage. Always either leave the current cassette on stage or transfer another cassette.
If there is any uncertainty about the job executing correctly, always use the END command followed by the cassette used during the exposure. If execution of a sequence fails the software will consider the sequence as executed and move on to the next part of the SDF file. If this is an END command that changes to another cassette the cassettte will simply be exchanged without giving the user a chance to correct the error and try again.
Jobdeck File
One or more Jobdeck files (.jdf) is needed for an exposure. As a minimum the JDF file must contain information of what pattern to write and where to write it. It can also be used to specify alignment information. The filename is limited to 24 characters, no capital letters and no spaces. The JDF file has a common block and one or more layer blocks. The layer blocks can be used to define several patterns and several writing conditions in a single file. A simple example of a JDF with a single layer block is given below.
;thope220101.jdf example file
JOB/W 'MYWAFER', 4
PATH DRF5M
ARRAY (-750,3,500)/(250,2,750)
ASSIGN P(1) -> (*,*)
SKIP (2,2)
AEND
PEND
LAYER 1
P(1) 'template1.v30'(0,0)
STDCUR 2.2
END
JDF common block
JOB [/W] [‘name’], d1 [, d2]
The common block starts with a JOB command. The /W switch indicates a round wafer slot, if omitted the system expects a square mask slot. Almost all cassettes at DTU Nanolab are wafer cassettes and thus require the /W switch for the pattern to be exposed at the correct slot coordinates. The name is optional and limited to 9 alphanumeric characters in uppercase. Following the optional name is the slot size designation d1, unit is inches. In the example it is a 4” wafer. A second dimension d2 can be added to constrain chip placement from the ARRAY command within that dimension, see below for an example of this.
Be aware that the chip cassettes at DTU Nanolab are converted 3” wafer cassettes and hence the correct header for an exposure on these will be JOB /W ‘name’, 3.
PATH [‘name’]
The PATH command defines the start of a writing path and defines the initial calibration routine carried out before pattern exposure as well as the cyclic calibration routine carried out during exposure. The PATH name refers to the exact name of the PATH as it exists on the system, the full list of available paths is available here.
DRF5M: INITIAL: CURRNT,HEIMAP CYCLIC: CURRNT,DRIFT CYCLE: 5M,F
Calibration routines under the “INITIAL” part of the path will be carried out prior to exposure. Normally a number of pre calibration routines are carried out manually by the operator. This makes it possible for the operator to gauge the result and act if something is out of the ordinary. In principle all of these routines can be run automatically if placed under the “INITIAL” part of a path. If the system encounters an error during this the entire sequence is however aborted. Please consult with the E-beam staff if you have specific requests in this regard.
Calibration routines under the “CYCLIC” part of the path are executed during pattern writing at the interval indicated in the path. Normally only beam current (CURRNT) and stage drift (DRIFT) is monitored during pattern writing. As an example of a path definition let us look at ‘DRF5M’.
During initial calibration only beam current and substrate height is measured. Thus all other initial calibration routines should be carried out manually with this path. During exposure beam current and stage drift is monitored and corrected every five minutes. The ‘F’ switch indicate that cyclic calibration will interrupt the current chip exposure upon completion of the current exposure field at the interval mark. If the ‘F’ switch is omitted cyclic calibration will wait for completion of the entire current chip before cyclic calibration is carried out.
PEND
The PEND command is required and is used to designate the end of a PATH.
ARRAY (x, nx, dx)/(y, ny, dy)
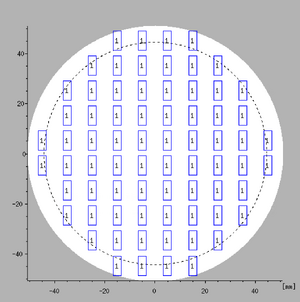
Every pattern must be part of an array to be exposed. Even in the simplest case where a single instance is to be written in the center of a substrate it must be defined with the ARRAY command. The ARRAY command essentially defines array points, i.e. points where pattern elements can be assigned to. Hence it works in conjunction with the ASSIGN command. The ARRAY command has six variables where x and y are the center coordinate of element (1,1) in µm, nx and ny are the number of array elements in x and y and dx and dy are the x- and y-axis pitches in µm. Notice that element (1,1) is the top left element of the array, additional x-axis elements are placed to the right of it and additional y-axis elements are placed below it. The JDF example above defines a 3 x 2 array with element (1,1) centered at (-750,250), x-axis pitch is 500 µm and y-axis pitch is 750 µm, as illustrated in Figure 3.
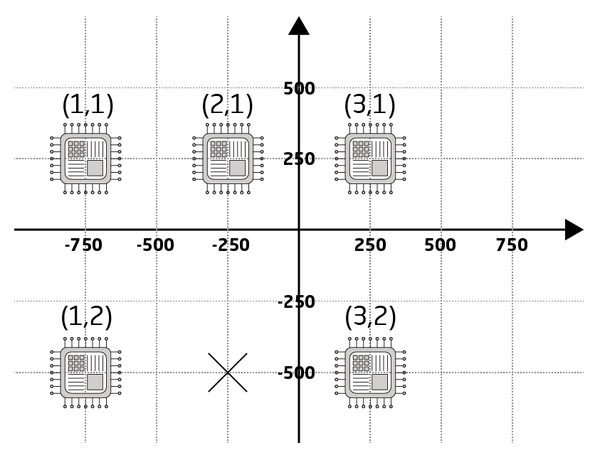
|
|
Visualization of the array defined using the ARRAY, ASSIGN and SKIP commands in the example JDF. Illustration: Thomas Pedersen. |
ASSIGN P(i) -> ((j,k),t)
The ASSIGN command is used to assign patterns to array points. It assigns the pattern element “P(i)” on the left side of the arrow to the array elements listed on the right hand side “(j,k)”. It also allows designation of a dose modulation table “t” to be used. In the example pattern P(1) is assigned to all elements of the array in both axis using the “*” character. No modulation table is referenced and hence the pattern is written without dose modulation. Notice that a ranges of elements can be assigned by writing a range in the assignment parenthesis. For instance, ASSIGN P(1) -> (3-5,*) will asign pattern 1 to all rows if columns 3, 4 and 5.
Also notice that if an array element is assigned several times, it is the last assignment that will be used.
The ASSIGN command can be used to build arrays of arrays to a depth of 9 sub arrays.
SKIP (j,k)
The skip command can be used to unassign an array element if it is to be left blank. In the example element (2,2) is unassigned and hence no pattern will be placed in this array element.
AEND
Array definition must end with an AEND command.
PEND
Path definition must end with a PEND command.
LAYER [number]
The layer command allows the user to define different writing conditions to different patterns by dividing them into different layers. Please notice that this “layer” has nothing to do with the layers in the user GDS pattern file. Each layer is called from the SDF using the JDF command. In the example here, LAYER 1 is defined in the JDF and in the SDF this is called by the command “JDF ‘thope220101’,1”, where the last number defines the JDF layer reference. In many cases users will just have a single layer. It is however possible to define several layers with different exposure conditions such as dose or current.
P(i) [‘patternfile.v30’][(dx,dy)]
In the layer section the P(i) command is used to define a pattern reference to a specific V30 pattern file to be written. Multiple patterns can be defined in a single layer. The pattern file must be placed in /home/eb0/jeoleb/pattern/danchipv30 to be read by the system and must be a V30 file. A position shift can be applied by specifying dx and dy, unit is µm. It is best practice to offset patterns in the ARRAY definition in the JDF or using the OFFSET command in SDF but it is possible to offset individual patterns in the P(i) definition. In the example P(1) is defined to be “template1.v30” with zero shift applied. If no pattern shift is applied the parenthesis can be omitted.
STDCUR [beam current]
Definition of beam current in nA for writing time estimation and dwell/shot time calculation. It has no effect on the actual exposure. Actual exposure beam current is controlled with the CALPRM command in SDF. It is however still important to use the STDCUR command correctly. The system has a 100 MHz scanner and thus the minimum shot time is 10 ns. During file compilation the system will calculate the minimum shot time based on the current defined in STDCUR. If STDCUR is set lower than the actual beam current measured during exposure there is a risk of violating the 10 ns limit and hence exposure will fail. Good practice is to set STDCUR to the expected beam current (based on selected CALPRM) and add 10% as a safety buffer. In the example it is expected that the beam current is 2 nA (from CALPRM 2na_ap4) and hence STDCUR is defined as 2.2 nA.
END
The JDF is terminated with a END command.
Alignment exposure
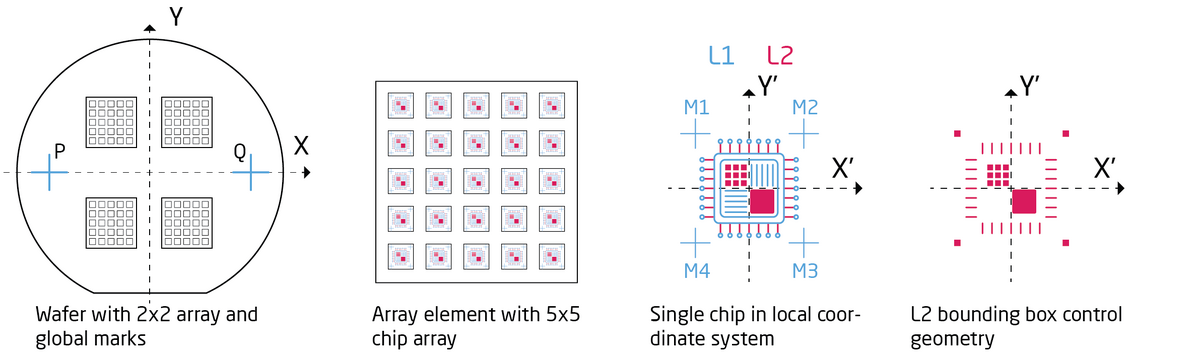
As described in the pattern preparation section there is two different strategies for pattern alignment. One can expose a wafer scale layout based on two global alignment marks or one can do chip exposures where each chip is individually aligned to an individual chip mark, as illustrated below. In both cases the SDF and JDF files have to be expanded with additional features to call and define the alignment procedure. Below is two examples, one for global alignment and one for chip alignment. Only new commands relative to the above example will be commented.
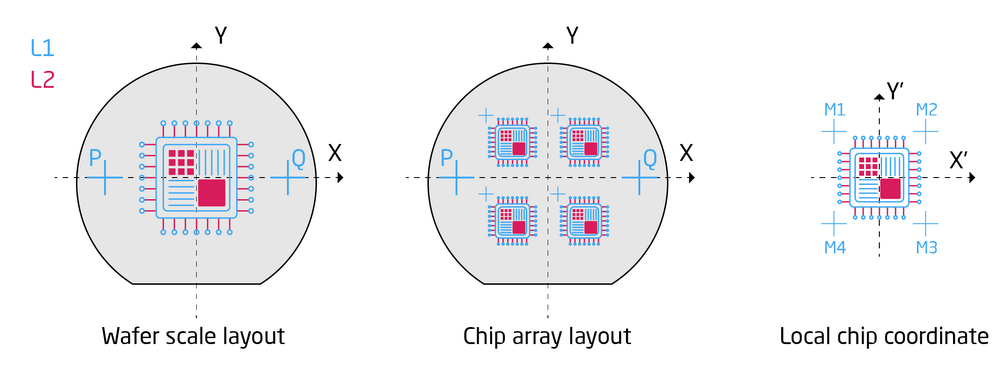
|
|
Illustration of wafer scale pattern alignment and chip array alignment for two designs, L1 and L2. The goal is to align the L2 pattern to the L1 pattern. |
Global alignment
The following files set up a single pattern for exposure with global alignment. To execute global alignment a few extra commands are used compared to the previous example. Only new commands are explained below.
Global alignment SDF
MAGAZIN 'ELELOP' #8 % 4A JDF 'elelop230401',1 ACC 100 CALPRM '6na_ap5' DEFMODE 2 GLMDET A CHIPAL 0 HSWITCH ON,OFF RESIST 152 SHOT A,32 OFFSET(-150,233) END 8
Compared to the first print exposure the SDF file has three extra commands, GLMDET, CHIPAL and HSWITCH. Additionally, the OFFSET command serves a somewhat different purpose now as explained below.
GLMDET [mode]
GLMDET (GLobal Mark DETect) has four modes
- A - for automatic alignment
- S - for semi automatic alignment
- M - for manual alignment
- C - for cancel, i.e. no alignment
In automatic mode the system will execute the SETWFR subprogram to find the global alignment marks (P and Q) by beam scans. SETWFR will execute mark detection at the P and Q mark locations defined in the JDF file. If alignment fails the system will continue to the next sequence in the SDF file. In semi automatic mode the system will carry out the same alignment procedure as automatic mode but if the mark scan fails the system will ask the user to use SEM mode to manually find the mark. When the mark is found in SEM mode the user can choose to designate the position as the true mark position or request another beam scan at the found location. Thus, semi automatic is somewhat more versatile at it allows the user to manually find the mark upon failure. Manual alignment allows the user to use SEM mode and manually find and identify the location of the center of the mark. Cancel mode skips global alignment. In the example above automatic alignment is used.
CHIPAL [mode]
CHIPAL has six modes and is explained in the section below on chip alignment. Even though chip alignment is not used during global alignment it is necessary to include the command and set it to either 0, V1 or V4, as explained below. In the example above CHIPAL is disregarded by setting it to 0.
HSWITCH [swg,swc]
HSWITHC specifies the substrate height detection mode for aligned exposure. It takes two parameters swg and swc which are merely two ON/OFF switches for global mark and chip marks, respectively. If swg is set to ON the system will carry out substrate height measurement at the global mark positions. If swc is set to ON the system will carry out substrate height measurement at the chip mark positions. Thus this is a convenient way to have the system measure substrate height and adjust beam focus to chip placement positions. In the example above the global mark positions are used for height detection of the substrate.
OFFSET[x,y]
For aligned exposures the OFFSET command must be used to state the offset of the P mark as determined by the optical pre-aligner. This offset will be applied to the entire content of the JDF file, i.e. the offset is used to shift the mark scan detection position. In the example above the pre-aligner was used to determine the P mark is shifted -150 µm in x and 233 µm in y relative to the center of the exposure/cassette slot.
Global alignment JDF
JOB/W 'ELELOP',4
GLMPOS P =(-36000,0), Q=(36000,0)
GLMP 2, 450,0,0
PATH DRF5M
ARRAY (0,1,0)/(0,1,0) ;
ASSIGN P(1) -> (*,*) ;
AEND
PEND
LAYER 1
P(1) 'elelop_GIST_L2.v30'
SPPRM 4.0,,,,1.0,1
STDCUR 6.6
OBJAPT 5
END
The JDF file has two additional commands, GLMPOS and GLMP.
GLMPOS [P=(x1,y1), Q=(x2,y2)]
GLMPPOS defines the global alignment mark coordinates in the substrate coordinate system and hence where SETWFR will perform beam scans for mark detection, unit is µm. In the example file the global marks are located at x = ±36000 µm and y = 0.
GLMP [w,l,t,f]
GLMP allows the user to add dimension information about the alignment mark and toggle between a cross and an L mark. The first parameter, w, defines the width of the mark, unit is µm. The second parameter, l, defines the length of the mark, unit is µm. For a cross this is the length of a single leg of the cross. The third parameter, t allows the user to toggle between a cross (0) and an L (1). The last parameter, f, defines the rotation of the mark in case an L mark is used. It is most advisable to use a cross, i.e. keep the last two parameters at 0. In the example above a 2 µm line width cross with a length of 450 µm is used.
Chip alignment
Chip alignment requires a global alignment to be made first to establish the wafer coordinate system. Hence a global alignment using GLMDET is used initially as in the example above. To further illustrate chip alignment we will look at a particular layout shown below. The layout also illustrates a commonly used feature on the JEOL system to create arrays of arrays. In the layout below there is main 2x2 array and into each of these is a 5x5 subarray. Each element of the subarray is a single chip with four chip alignment marks. Notice that array placement is given in the substrate coordinate system and so is the global mark positions. Chip alignment marks (M1-M4) are however given in the local chip coordinate system. In the example files below we assume L1 is already defined on the substrate and we wish to align L2 to it.
Since a pattern (V30 file) is placed at the center of the bounding box it is essential to control the bounding box of the chip design. The design in this case appears symmetric around (0,0) but in order to force it to be symmetric it is common to place corner marks at equidistance points from (0,0). The corner marks can be 1 nm boxes that will not show up in the resist when developed.
Chip alignment SDF
MAGAZIN 'THOPE' #8 %4C JDF 'thope230126',1 ACC 100 CALPRM '6na_ap5' DEFMODE 2 GLMDET S CHIPAL 1 HSWITCH OFF,ON RESIST 250 SHOT A,6 OFFSET(-44,-139) END 8
CHIPAL [mode]
CHIPAL has six modes
- 0 - Cancels chip alignment
- S - SEM mode. The user is prompted to use SEM mode to manually find M1
- 1 - One mark is used for position correction
- 4 - Four marks are used for position, rotation and gain correction
- V1 - Virtual mode 1. A single mark position is used for height detection of the substrate, no position correction
- V4 - Virtual mode 4. Four mark positions are used for height detection of the substrate, no position correction
In addition to position correction mode 1 and 4 also detects substrate height. The virtual modes are only used to detect substrate height since no mark detection is done. Mode S obviously very time consuming for a high number of chips.
If set up properly on good quality marks mode 1 or mode 4 chip alignment can usually execute in about 1-2 seconds per mark. The time estimate at compilation will account for the time spend on chip alignment at the current settings of the CHIPAL subprogram.
Chip alignment JDF
In addition to the use of CHMPOS for chip mark position definition the example below illustrate making arrays of an array. The first array is set up as a 2x2 array assigning array A1. A1 is defined below as 1:, since 1 is defined as an array it can be referenced as A1. A then defines a 5x5 array assigning pattern P(1) to each element. The chip mark position command must be used in the same array that assigns the corresponding pattern.
JOB/W 'THOPE',4 ; 4inch wafer
GLMPOS P=(-30000,0),Q=(30000,0)
PATH FT01
ARRAY (-15000,2,30000)/(10000,2,20000)
ASSIGN A(1) -> ((1,1))
1: ARRAY (-4000,5,2000)/(4000,5,2000)
CHMPOS M1=(-450,450),M2=(450,450),M3=(450,-450),M4=(-450,-450)
ASSIGN P(1) -> (*,*)
AEND
PEND
LAYER 1
P(1) 'thope230126.v30'
SPPRM 4.0,,,,1.0,1
STDCUR 6
END
CHMPOS [M1=(x1,y1){,M2=(x2,y2),M3=(x3,y3),M4=(x4,y4)}]
CHMPOS defines the chip alignment marks in the local chip coordinate system, unit is µm. M1 is mandatory while M2-M4 are optional in one mark mode. In four mark mode all marks must be defined. The order of the marks is important, M1 must be top left with M2-M4 placed clockwise around the center. In the given example the four marks are placed symmetrically at ±450 µm in x and ±450 µm in y.
It V1 mode it is customary to set M1 = (0,0) such that substrate height is detected at the center of the chip. In this way V1 mode can be used to exactly read out substrate height where the chip pattern will be written.
Beam current and condition files
The beam current can in principle be changed in very fine steps, it however requires recalibration of the Dynamic Focus and Dynamic Stigmation table. Hence, only a limited number of beam currents are available. The available beam currents and condition file name are listed below.
| Beam current [nA] | Aperture | Condition file |
|---|---|---|
| 0.12 | 4 | 0.12na_ap4 |
| 0.16 | 4 | 0.16na_ap4 |
| 0.22 | 4 | 0.22na_ap4 |
| 0.4 | 4 | 0.4na_ap4 |
| 0.5 | 4 | 0.5na_ap4 |
| 0.8 | 4 | 0.8na_ap4 |
| 1.4 | 4 | 1.4na_ap4 |
| 1.6 | 4 | 1.6na_ap4 |
| 2 | 4 | 2na_ap4 |
| 2.7 | 4 | 2.7na_ap4 |
| 3.8 | 5 | 3.8na_ap5 |
| 4 | 4 | 4na_ap4 |
| 5 | 5 | 5na_ap5 |
| 6 | 5 | 6na_ap5 |
| 10 | 6 | 10na_ap6 |
| 12 | 5 | 12na_ap5 |
| 14 | 8 | 14na_ap8 |
| 19 | 7 | 19na_ap7 |
| 21 | 7 | 21na_ap7 |
| 22 | 7 | 22na_ap7 |
| 25 | 7 | 25na_ap7 |
| 27 | 7 | 27na_ap7 |
| 29 | 7 | 29na_ap7 |
| 30 | 8 | 30na_ap8 |
| 36 | 8 | 36na_ap8 |
| 41 | 8 | 41na_ap8 |
| 44 | 8 | 44na_ap8 |
| 54 | 7 | 54na_ap7 |
| 60 | 8 | 60na_ap8 |