Specific Process Knowledge/Lithography/EBeamLithography/Dose Testing: Difference between revisions
No edit summary |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
'''Feedback to this page''': '''[mailto:labadviser@nanolab.dtu.dk?Subject=Feed%20back%20from%20page%20http://labadviser.nanolab.dtu.dk/index.php/Specific_Process_Knowledge/Lithography/EBeamLithography/Dose Testing click here]''' | |||
Content and illustration by Thomas Pedersen, DTU Nanolab unless otherwise noted. | |||
=Introduction= | =Introduction= | ||
In E-beam lithography it is often necesarry to do a dose test in order to get the required result. In a dose test one will expose critical parts of a pattern with various doses and determine the best dose by SEM analysis of the final pattern. There are several ways to set up a dose test array on the JEOL system, in this section we will describe four different setups, each with their own benefits and drawbacks. | In E-beam lithography it is often necesarry to do a dose test in order to get the required result. In a dose test one will expose critical parts of a pattern with various doses and determine the best dose by SEM analysis of the final pattern. There are several ways to set up a dose test array on the JEOL system, in this section we will describe four different setups, each with their own benefits and drawbacks. | ||
| Line 147: | Line 151: | ||
|- | |- | ||
| colspan="1" style="text-align: center;| | | colspan="1" style="text-align: center;| | ||
Example frome ChipPlace definine a 10 x 2 element dose matrix with relative doses from 1 to 2, ready for export to V30 | Example frome ChipPlace definine a 10 x 2 element dose matrix with relative doses from 1 to 2, ready for export to V30. | ||
|} | |} | ||
=Scripted expansion of a PEC modulation table= | =Scripted expansion of a PEC modulation table= | ||
The last method is basically the same as the second method above but with support for PEC modulation. This is achieved by expanding the dose modulation table into a set of tables, one for each dose of the dose test. Each modulation table is a scaled version of the initial modulation table, scaled simply by the dose test scaling between each dose. The scaling is done through a Matlab script which can be found | The last method is basically the same as the second method above but with support for PEC modulation. This is achieved by expanding the dose modulation table into a set of tables, one for each dose of the dose test. Each modulation table is a scaled version of the initial modulation table, scaled simply by the dose test scaling between each dose. The scaling is done through a Matlab script which can be found below. Since Labadviser does not allow upload of .m files, simply copy the script from below and save as a .m file. | ||
Benefits: | Benefits: | ||
| Line 167: | Line 171: | ||
| | | | ||
<pre> | <pre> | ||
clearvars | |||
%Modulation table modding for dose test of PEC'ed V30's, see Labadviser for further information | %Modulation table modding for dose test of PEC'ed V30's, see Labadviser for further information | ||
%https://labadviser.nanolab.dtu.dk/index.php?title=Specific_Process_Knowledge/Lithography/EBeamLithography/Dose_Testing | %https://labadviser.nanolab.dtu.dk/index.php?title=Specific_Process_Knowledge/Lithography/EBeamLithography/Dose_Testing | ||
%THOPE 2023-11- | %THOPE 2023-11-13 | ||
%Input variables for modulation table | %Input variables for modulation table | ||
inputJDI = 'C:\Users\thope\Desktop\ | inputJDI = 'C:\Users\thope\Desktop\thope231113\withcross.jdi'; | ||
outputJDI = ' | outputJDI = 'C:\Users\thope\Desktop\thope231113\thope231113.jdi'; | ||
m = 9; %number of doses | m = 9; %number of doses | ||
modulation = | modulation = 5; %modulation between instances in percent, must be positive | ||
%shot time calculation | |||
Base = 220; %Base dose, only for indication, not used for modulation | |||
I = 2e-9; %beam current | |||
pitch = 12; %steps of 0.25 nm | |||
dp = pitch*0.25*1e-9; %beam step in m | |||
%Base dose calculation | |||
Doses = (0:1:m-1); | |||
Doses = (Doses*modulation/100+1)*Base; | |||
%Read file | %Read file | ||
| Line 201: | Line 215: | ||
[class(i),D(1,i)] = readMOD(A,k1(i),k2(i)); | [class(i),D(1,i)] = readMOD(A,k1(i),k2(i)); | ||
end | end | ||
%Calculate tables | %Calculate tables | ||
D(1,:) = 1 + D(1,:)./100 ; %convert from MOD to percent | D(1,:) = 1 + D(1,:)./100 ; %convert from MOD to percent | ||
for i = 2: | for i = 2:m | ||
D(i,:) = D(1,:)*(1+modulation*(i-1)/100); | D(i,:) = D(1,:)*(1+modulation*(i-1)/100); | ||
end | end | ||
D = round(D,3); %round to 1 decimal | D = round(D,3); %round to 1 decimal | ||
for i = 1: | for i = 1:m | ||
D(i,:) = 100*(D(i,:)-1); %convert to MOD from percent | D(i,:) = 100*(D(i,:)-1); %convert to MOD from percent | ||
end | end | ||
%Write modulated tables to file | %Write modulated tables to file | ||
| Line 232: | Line 248: | ||
end | end | ||
fclose(fileID); | fclose(fileID); | ||
%Modulation table range | |||
range = zeros(m,2); | |||
for i = 1:m | |||
range(i,1) = D(i,1); %minimum dose | |||
range(i,2) = D(i,end); %maximum dose | |||
end | |||
range = (range/100+1)*Base; | |||
%Frequency range | |||
F = zeros(m,2); | |||
for i = 1:m | |||
F(i,1) = I./(range(i,1)*dp^2); %minimum dose | |||
F(i,2) = I./(range(i,2)*dp^2); %maximum dose | |||
end | |||
F = F/1e4; | |||
%Display output | |||
disp(newline) | |||
disp('Modulation table | Base dose [µC/cm2] | Modulation range [µC/cm2] | Frequency range [MHz]') | |||
for i = 1:m | |||
disp(['MOD' num2str(i) ' | ' num2str(Doses(i)) ' | ' num2str(range(i,1),'%.0f')... | |||
' - ' num2str(range(i,2),'%.0f') ' | ' num2str(F(i,1),'%.0f') ' - ' num2str(F(i,2),'%.0f')]) | |||
end | |||
function [a,b] = readMOD(str,k1,k2) | function [a,b] = readMOD(str,k1,k2) | ||
Latest revision as of 08:03, 14 November 2023
Feedback to this page: Testing click here
Content and illustration by Thomas Pedersen, DTU Nanolab unless otherwise noted.
Introduction
In E-beam lithography it is often necesarry to do a dose test in order to get the required result. In a dose test one will expose critical parts of a pattern with various doses and determine the best dose by SEM analysis of the final pattern. There are several ways to set up a dose test array on the JEOL system, in this section we will describe four different setups, each with their own benefits and drawbacks.
Simple SDF sequencing
The most straightforward method is to create one SDF sequence per dose and simply vary the dose with the RESIST command in the SDF as illustrated below. Each instance must of course be offset with the OFFSET command such that they are not exposed on top of each other. The pattern information in the referenced JDF can contain a PEC modulation table with this method and hence it is suited for proximity corrected designs.
Benefits:
- Easy to set up
- Supports PEC modulation
Drawbacks:
- The system will perform initial calibration according to the PATH defined in the JDF for each sequence/dose. This will add several minutes of execution time to each sequence and for a large dose matrix it can cost a lot of additional time.
;SDF example
MAGAZIN 'DOSES'
;--------Dose 1--------
#1
%4D
JDF 'dosetest',1
ACC 100
CALPRM '6na_ap5'
DEFMODE 2
RESIST 200
SHOT A,24
OFFSET(-100,0)
;--------Dose 2--------
#1
%4D
JDF 'dosetest',1
ACC 100
CALPRM '6na_ap5'
DEFMODE 2
RESIST 220
SHOT A,24
OFFSET(0,0)
;--------Dose 3--------
#1
%4D
JDF 'dosetest',1
ACC 100
CALPRM '6na_ap5'
DEFMODE 2
RESIST 240
SHOT A,24
OFFSET(100,0)
END
Dose modulation without PEC
The MODULAT command can be used to modulate the base dose for a pattern and hence it can be used to generate a dose matrix. In this setup the SDF will contain a base dose defined by the RESIST command, this dose will then be modulated for different pattern instances defined in the JDF. In this setup the SDF will only contain a single sequence as below.
;SDF example
MAGAZIN 'DOSES'
#1
%4B
JDF 'dosetest',1
ACC 100
CALPRM '6na_ap5'
DEFMODE 2
RESIST 200
SHOT A,20
OFFSET(0,0)
END 1
The arrray is setup with the ARRAY command in the JDF and at the end of the ASSIGN command a dose modulation can be applied. In this way each instance assignment can contain a different modulation. In the example below the pattern is instanced in a 10 x 1 matrix, each element has its own modulation table; SHOT1 to SHOT10. The definition of each modulation is stated at the end of the layer definition.
The MODULAT command takes two parameters as MODULAT(r,v), where r is the shot rank and v is the shot time modulation in %. The shot rank is defined during export from Beamer. For a simple design as used in this example that is not proximity corrected all elements of the pattern will be in shot rank 0. The shot time modulation is a simple percentage increase to the base dose defined by the RESIST command in the SDF. The modulation table in this example will thus expose with a base dose of 200 µC/cm2 in element (1,1) and a dose of 200 µC/cm2 + 45% = 290 µC/cm2 in element (10,1). The resulting pattern and modulation is visualised below.
Benefits:
- Easy to set up
Drawbacks:
- Does not support PEC modulation since we use the MODULAT option to define the dose variation
;JDF example
JOB/W 'DOSES',4
PATH DRF5M
ARRAY (50,10,50)/(50,1,0)
ASSIGN P(1)->((1,1),SHOT1)
ASSIGN P(1)->((2,1),SHOT2)
ASSIGN P(1)->((3,1),SHOT3)
ASSIGN P(1)->((4,1),SHOT4)
ASSIGN P(1)->((5,1),SHOT5)
ASSIGN P(1)->((6,1),SHOT6)
ASSIGN P(1)->((7,1),SHOT7)
ASSIGN P(1)->((8,1),SHOT8)
ASSIGN P(1)->((9,1),SHOT9)
ASSIGN P(1)->((10,1),SHOT10)
AEND
PEND
LAYER 1
P(1) 'dtu_logo_um.v30'
SPPRM 4.0,,,,1.0,1
STDCUR 6.6 ;nA
SHOT1: MODULAT (( 0,0))
SHOT2: MODULAT (( 0,5))
SHOT3: MODULAT (( 0,10))
SHOT4: MODULAT (( 0,15))
SHOT5: MODULAT (( 0,20))
SHOT6: MODULAT (( 0,25))
SHOT7: MODULAT (( 0,30))
SHOT8: MODULAT (( 0,35))
SHOT9: MODULAT (( 0,40))
SHOT10: MODULAT (( 0,45))
END
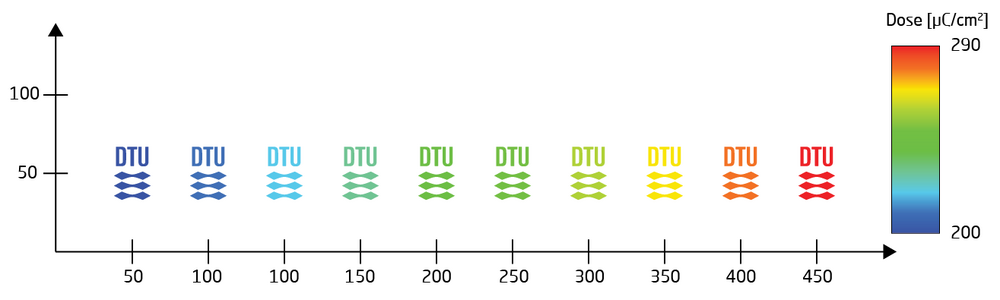
|
|
Resulting setup from the example job. The pattern (DTU logo) is instanced 10 times with a pitch of 50 µm. The dose is modulated between 200 and 290 µC/cm2. |
Using ChipPlace in Beamer - recommended for most cases
ChipPlace is part of Beamer and offers a graphical user interface to layout exposure jobs. In ChipPlace one can create arrays of pattern elements and assign doses to each pattern element. The output is a single design file and a single modulation table. The design file will contain all pattern elements and can be exported to V30. The modulation table is exported as JDI and will contain any PEC modulation applied to the design as well as modulation from the chosen dose range. Please refer to the Beamer page for a guide on how to set this up. This method supports PEC modulation and is recommendable for most users.
Benefits:
- Collects all pattern elements in a single V30 and a single JDI
- Supports PEC modulation
- Can easily generate dose labels written next to the array elements such that the user does not have to remember how the matrix is set up
Drawbacks
- A bit more time consuming to set up
- Can generate very large V30 files since the design is repeated once per dose. For small designs this is no problem, for large (1 GB) designs this becomes a problem.
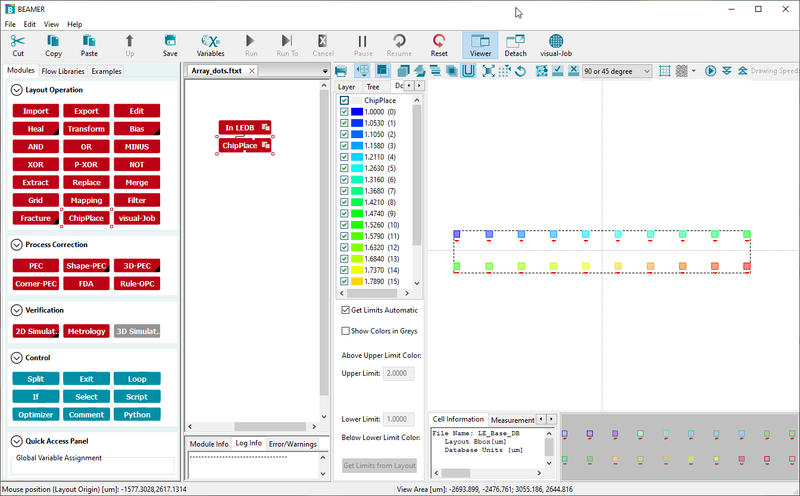
|
|
Example frome ChipPlace definine a 10 x 2 element dose matrix with relative doses from 1 to 2, ready for export to V30. |
Scripted expansion of a PEC modulation table
The last method is basically the same as the second method above but with support for PEC modulation. This is achieved by expanding the dose modulation table into a set of tables, one for each dose of the dose test. Each modulation table is a scaled version of the initial modulation table, scaled simply by the dose test scaling between each dose. The scaling is done through a Matlab script which can be found below. Since Labadviser does not allow upload of .m files, simply copy the script from below and save as a .m file.
Benefits:
- Supports PEC modulation
- Faster execution since it is a single SDF sequence
- Works with large V30 files
Drawbacks:
- A bit tricky for first time use
| Matlab script for modulation table expansion |
|---|
clearvars
%Modulation table modding for dose test of PEC'ed V30's, see Labadviser for further information
%https://labadviser.nanolab.dtu.dk/index.php?title=Specific_Process_Knowledge/Lithography/EBeamLithography/Dose_Testing
%THOPE 2023-11-13
%Input variables for modulation table
inputJDI = 'C:\Users\thope\Desktop\thope231113\withcross.jdi';
outputJDI = 'C:\Users\thope\Desktop\thope231113\thope231113.jdi';
m = 9; %number of doses
modulation = 5; %modulation between instances in percent, must be positive
%shot time calculation
Base = 220; %Base dose, only for indication, not used for modulation
I = 2e-9; %beam current
pitch = 12; %steps of 0.25 nm
dp = pitch*0.25*1e-9; %beam step in m
%Base dose calculation
Doses = (0:1:m-1);
Doses = (Doses*modulation/100+1)*Base;
%Read file
fileID=fopen(inputJDI,'r');
A = fscanf(fileID,'%c');
fclose(fileID);
%Find start of modulation table by searching for '('
k1 = strfind(A, '(');
k1 = k1(2:end-1);
%Find end of modulation table by searching for ')'
k2 = strfind(A, ')');
k2 = k2(1:end-2);
n = numel(k1);
%Display input table
disp('Input table:')
disp(A(1:k2(end)+1))
%Define zero matrix
class = zeros(n,1);
D = zeros(m,n);
%Read base modulation
for i = 1:n
[class(i),D(1,i)] = readMOD(A,k1(i),k2(i));
end
%Calculate tables
D(1,:) = 1 + D(1,:)./100 ; %convert from MOD to percent
for i = 2:m
D(i,:) = D(1,:)*(1+modulation*(i-1)/100);
end
D = round(D,3); %round to 1 decimal
for i = 1:m
D(i,:) = 100*(D(i,:)-1); %convert to MOD from percent
end
%Write modulated tables to file
fileID = fopen(outputJDI,'w');
for i = 1:m
fprintf(fileID,['MOD' num2str(i) ': MODULAT (']);
for k = 1:size(D,2)
if k==1
fprintf(fileID,['( ' num2str(k-1) ', ' num2str(D(i,k)) ' )']);
else
if mod(k,3)
fprintf(fileID,[' , ( ' num2str(k-1) ', ' num2str(D(i,k)) ' )']);
else
fprintf(fileID,[' , ( ' num2str(k-1) ', ' num2str(D(i,k)) ' )']);
fprintf(fileID,['\n' '- ']);
end
end
end
fprintf(fileID,')');
fprintf(fileID,['\n\n']);
end
fclose(fileID);
%Modulation table range
range = zeros(m,2);
for i = 1:m
range(i,1) = D(i,1); %minimum dose
range(i,2) = D(i,end); %maximum dose
end
range = (range/100+1)*Base;
%Frequency range
F = zeros(m,2);
for i = 1:m
F(i,1) = I./(range(i,1)*dp^2); %minimum dose
F(i,2) = I./(range(i,2)*dp^2); %maximum dose
end
F = F/1e4;
%Display output
disp(newline)
disp('Modulation table | Base dose [µC/cm2] | Modulation range [µC/cm2] | Frequency range [MHz]')
for i = 1:m
disp(['MOD' num2str(i) ' | ' num2str(Doses(i)) ' | ' num2str(range(i,1),'%.0f')...
' - ' num2str(range(i,2),'%.0f') ' | ' num2str(F(i,1),'%.0f') ' - ' num2str(F(i,2),'%.0f')])
end
function [a,b] = readMOD(str,k1,k2)
str = str(k1:k2);
x = regexp(str,',');
a = str2double(str(2:x-1));
b = str2double(str(x+1:end-1));
end
|
The number of doses for dose testing is controlled by the parameter m while the increase in dose between doses is defined by modulation, it is a percentwise change relative to the base dose. For instance, if one wants to do a dose test with 200 µC/cm2 as base dose in steps of 10 µC/cm2 one would set modulation = 5.
The output is a single JDI file containing all modulation tables named MODx, where x is a running number from 1 to m. The array and reference to each modulation table can then be set up in the JDF file as seen below. It is convenient to use the include command @ to reference the JDI file itself rather than copy-paste the tables into the JDF.
| Example JDF with a dose test setup defined by the ARRAY command and reference to individual modulation tables. |
|---|
JOB/W 'THOPE',4
PATH DRF5M
ARRAY (-4000,5,2000)/(0,1,0)
ASSIGN P(1) -> ((1,1),MOD1)
ASSIGN P(1) -> ((2,1),MOD2)
ASSIGN P(1) -> ((3,1),MOD3)
ASSIGN P(1) -> ((4,1),MOD4)
ASSIGN P(1) -> ((5,1),MOD5)
AEND
PEND
LAYER 1
P(1) 'mypattern.v30'
SPPRM 4.0,,,,1.0,1
STDCUR 2.0 ;nA
@'thopeExpanded.jdi'
END
|
| Example JDI file as output by the Matlab script. |
|---|
MOD1: MODULAT (( 0, -23.6 ) , ( 1, -22.8 ) , ( 2, -22 ) - , ( 3, -21.2 ) , ( 4, -20.5 ) , ( 5, -19.7 ) - , ( 6, -18.9 ) , ( 7, -18 ) , ( 8, -17.2 ) - , ( 9, -16.4 ) , ( 10, -15.6 ) , ( 11, -14.7 ) - , ( 12, -13.9 ) , ( 13, -13 ) , ( 14, -12.1 ) - , ( 15, -11.3 ) , ( 16, -10.4 ) , ( 17, -9.5 ) - , ( 18, -8.6 ) , ( 19, -7.7 ) , ( 20, -6.7 ) - , ( 21, -5.8 ) , ( 22, -4.8 ) , ( 23, -3.9 ) - , ( 24, -2.9 ) , ( 25, -2 ) , ( 26, -1 ) - , ( 27, 0 ) , ( 28, 1 ) , ( 29, 2 ) - , ( 30, 3 ) , ( 31, 4.1 ) , ( 32, 5.1 ) - , ( 33, 6.2 ) , ( 34, 7.2 ) , ( 35, 8.3 ) - , ( 36, 9.4 ) , ( 37, 10.5 ) , ( 38, 11.6 ) - , ( 39, 12.7 ) , ( 40, 13.8 ) , ( 41, 14.9 ) - , ( 42, 16.1 ) , ( 43, 17.3 ) , ( 44, 18.4 ) - , ( 45, 19.6 ) , ( 46, 20.8 ) , ( 47, 22 ) - , ( 48, 23.2 ) , ( 49, 24.5 ) , ( 50, 25.7 ) - , ( 51, 27 ) , ( 52, 28.2 ) , ( 53, 29.5 ) - , ( 54, 30.8 ) , ( 55, 32.1 ) , ( 56, 33.5 ) - , ( 57, 34.8 ) , ( 58, 36.1 ) , ( 59, 37.5 ) - , ( 60, 38.9 ) , ( 61, 40.3 ) , ( 62, 41.7 ) - , ( 63, 43.1 ) , ( 64, 44.5 ) , ( 65, 46 ) - , ( 66, 47.4 ) , ( 67, 48.9 ) , ( 68, 50.4 ) - ) MOD2: MODULAT (( 0, -16 ) , ( 1, -15.1 ) , ( 2, -14.2 ) - , ( 3, -13.3 ) , ( 4, -12.5 ) , ( 5, -11.7 ) - , ( 6, -10.8 ) , ( 7, -9.8 ) , ( 8, -8.9 ) - , ( 9, -8 ) , ( 10, -7.2 ) , ( 11, -6.2 ) - , ( 12, -5.3 ) , ( 13, -4.3 ) , ( 14, -3.3 ) - , ( 15, -2.4 ) , ( 16, -1.4 ) , ( 17, -0.4 ) - , ( 18, 0.5 ) , ( 19, 1.5 ) , ( 20, 2.6 ) - , ( 21, 3.6 ) , ( 22, 4.7 ) , ( 23, 5.7 ) - , ( 24, 6.8 ) , ( 25, 7.8 ) , ( 26, 8.9 ) - , ( 27, 10 ) , ( 28, 11.1 ) , ( 29, 12.2 ) - , ( 30, 13.3 ) , ( 31, 14.5 ) , ( 32, 15.6 ) - , ( 33, 16.8 ) , ( 34, 17.9 ) , ( 35, 19.1 ) - , ( 36, 20.3 ) , ( 37, 21.6 ) , ( 38, 22.8 ) - , ( 39, 24 ) , ( 40, 25.2 ) , ( 41, 26.4 ) - , ( 42, 27.7 ) , ( 43, 29 ) , ( 44, 30.2 ) - , ( 45, 31.6 ) , ( 46, 32.9 ) , ( 47, 34.2 ) - , ( 48, 35.5 ) , ( 49, 37 ) , ( 50, 38.3 ) - , ( 51, 39.7 ) , ( 52, 41 ) , ( 53, 42.5 ) - , ( 54, 43.9 ) , ( 55, 45.3 ) , ( 56, 46.9 ) - , ( 57, 48.3 ) , ( 58, 49.7 ) , ( 59, 51.3 ) - , ( 60, 52.8 ) , ( 61, 54.3 ) , ( 62, 55.9 ) - , ( 63, 57.4 ) , ( 64, 59 ) , ( 65, 60.6 ) - , ( 66, 62.1 ) , ( 67, 63.8 ) , ( 68, 65.4 ) - ) MOD3: MODULAT (( 0, -8.3 ) , ( 1, -7.4 ) , ( 2, -6.4 ) - , ( 3, -5.4 ) , ( 4, -4.6 ) , ( 5, -3.6 ) - , ( 6, -2.7 ) , ( 7, -1.6 ) , ( 8, -0.6 ) - , ( 9, 0.3 ) , ( 10, 1.3 ) , ( 11, 2.4 ) - , ( 12, 3.3 ) , ( 13, 4.4 ) , ( 14, 5.5 ) - , ( 15, 6.4 ) , ( 16, 7.5 ) , ( 17, 8.6 ) - , ( 18, 9.7 ) , ( 19, 10.8 ) , ( 20, 12 ) - , ( 21, 13 ) , ( 22, 14.2 ) , ( 23, 15.3 ) - , ( 24, 16.5 ) , ( 25, 17.6 ) , ( 26, 18.8 ) - , ( 27, 20 ) , ( 28, 21.2 ) , ( 29, 22.4 ) - , ( 30, 23.6 ) , ( 31, 24.9 ) , ( 32, 26.1 ) - , ( 33, 27.4 ) , ( 34, 28.6 ) , ( 35, 30 ) - , ( 36, 31.3 ) , ( 37, 32.6 ) , ( 38, 33.9 ) - , ( 39, 35.2 ) , ( 40, 36.6 ) , ( 41, 37.9 ) - , ( 42, 39.3 ) , ( 43, 40.8 ) , ( 44, 42.1 ) - , ( 45, 43.5 ) , ( 46, 45 ) , ( 47, 46.4 ) - , ( 48, 47.8 ) , ( 49, 49.4 ) , ( 50, 50.8 ) - , ( 51, 52.4 ) , ( 52, 53.8 ) , ( 53, 55.4 ) - , ( 54, 57 ) , ( 55, 58.5 ) , ( 56, 60.2 ) - , ( 57, 61.8 ) , ( 58, 63.3 ) , ( 59, 65 ) - , ( 60, 66.7 ) , ( 61, 68.4 ) , ( 62, 70 ) - , ( 63, 71.7 ) , ( 64, 73.4 ) , ( 65, 75.2 ) - , ( 66, 76.9 ) , ( 67, 78.7 ) , ( 68, 80.5 ) - ) MOD4: MODULAT (( 0, -0.7 ) , ( 1, 0.4 ) , ( 2, 1.4 ) - , ( 3, 2.4 ) , ( 4, 3.4 ) , ( 5, 4.4 ) - , ( 6, 5.4 ) , ( 7, 6.6 ) , ( 8, 7.6 ) - , ( 9, 8.7 ) , ( 10, 9.7 ) , ( 11, 10.9 ) - , ( 12, 11.9 ) , ( 13, 13.1 ) , ( 14, 14.3 ) - , ( 15, 15.3 ) , ( 16, 16.5 ) , ( 17, 17.7 ) - , ( 18, 18.8 ) , ( 19, 20 ) , ( 20, 21.3 ) - , ( 21, 22.5 ) , ( 22, 23.8 ) , ( 23, 24.9 ) - , ( 24, 26.2 ) , ( 25, 27.4 ) , ( 26, 28.7 ) - , ( 27, 30 ) , ( 28, 31.3 ) , ( 29, 32.6 ) - , ( 30, 33.9 ) , ( 31, 35.3 ) , ( 32, 36.6 ) - , ( 33, 38.1 ) , ( 34, 39.4 ) , ( 35, 40.8 ) - , ( 36, 42.2 ) , ( 37, 43.7 ) , ( 38, 45.1 ) - , ( 39, 46.5 ) , ( 40, 47.9 ) , ( 41, 49.4 ) - , ( 42, 50.9 ) , ( 43, 52.5 ) , ( 44, 53.9 ) - , ( 45, 55.5 ) , ( 46, 57 ) , ( 47, 58.6 ) - , ( 48, 60.2 ) , ( 49, 61.9 ) , ( 50, 63.4 ) - , ( 51, 65.1 ) , ( 52, 66.7 ) , ( 53, 68.4 ) - , ( 54, 70 ) , ( 55, 71.7 ) , ( 56, 73.6 ) - , ( 57, 75.2 ) , ( 58, 76.9 ) , ( 59, 78.8 ) - , ( 60, 80.6 ) , ( 61, 82.4 ) , ( 62, 84.2 ) - , ( 63, 86 ) , ( 64, 87.9 ) , ( 65, 89.8 ) - , ( 66, 91.6 ) , ( 67, 93.6 ) , ( 68, 95.5 ) - ) MOD5: MODULAT (( 0, 7 ) , ( 1, 8.1 ) , ( 2, 9.2 ) - , ( 3, 10.3 ) , ( 4, 11.3 ) , ( 5, 12.4 ) - , ( 6, 13.5 ) , ( 7, 14.8 ) , ( 8, 15.9 ) - , ( 9, 17 ) , ( 10, 18.2 ) , ( 11, 19.4 ) - , ( 12, 20.5 ) , ( 13, 21.8 ) , ( 14, 23.1 ) - , ( 15, 24.2 ) , ( 16, 25.4 ) , ( 17, 26.7 ) - , ( 18, 28 ) , ( 19, 29.2 ) , ( 20, 30.6 ) - , ( 21, 31.9 ) , ( 22, 33.3 ) , ( 23, 34.5 ) - , ( 24, 35.9 ) , ( 25, 37.2 ) , ( 26, 38.6 ) - , ( 27, 40 ) , ( 28, 41.4 ) , ( 29, 42.8 ) - , ( 30, 44.2 ) , ( 31, 45.7 ) , ( 32, 47.1 ) - , ( 33, 48.7 ) , ( 34, 50.1 ) , ( 35, 51.6 ) - , ( 36, 53.2 ) , ( 37, 54.7 ) , ( 38, 56.2 ) - , ( 39, 57.8 ) , ( 40, 59.3 ) , ( 41, 60.9 ) - , ( 42, 62.5 ) , ( 43, 64.2 ) , ( 44, 65.8 ) - , ( 45, 67.4 ) , ( 46, 69.1 ) , ( 47, 70.8 ) - , ( 48, 72.5 ) , ( 49, 74.3 ) , ( 50, 76 ) - , ( 51, 77.8 ) , ( 52, 79.5 ) , ( 53, 81.3 ) - , ( 54, 83.1 ) , ( 55, 84.9 ) , ( 56, 86.9 ) - , ( 57, 88.7 ) , ( 58, 90.5 ) , ( 59, 92.5 ) - , ( 60, 94.5 ) , ( 61, 96.4 ) , ( 62, 98.4 ) - , ( 63, 100.3 ) , ( 64, 102.3 ) , ( 65, 104.4 ) - , ( 66, 106.4 ) , ( 67, 108.5 ) , ( 68, 110.6 ) - ) |
