Specific Process Knowledge/Characterization/XRD: Difference between revisions
| Line 36: | Line 36: | ||
Peak angles can be calculated by simple vector calculations; however, I have made a [[small MATLAB script]] calculating them for you. To use the script open MATLAB and call the program with two vectors as input. First vector should be your surface orientation, second input the plane you measure on. For a [1 1 1] substrate surface and a [3 1 1] plane and a [0 0 1] substrate surface and a [1 3 3] plane, where the measurement planes are dependent on the 2theta angle, the command looks like this: | Peak angles can be calculated by simple vector calculations; however, I have made a [[small MATLAB script]] calculating them for you. To use the script open MATLAB and call the program with two vectors as input. First vector should be your surface orientation, second input the plane you measure on. For a [1 1 1] substrate surface and a [3 1 1] plane and a [0 0 1] substrate surface and a [1 3 3] plane, where the measurement planes are dependent on the 2theta angle, the command looks like this: | ||
PoleFigureAngles([1 1 1; 0 0 1],[3 1 1; 1 3 3]) | {|style="margin:auto;" | ||
|PoleFigureAngles([1 1 1; 0 0 1],[3 1 1; 1 3 3]) | |||
|} | |||
It is also possible to give a path for a file exported from 3D Explore with contour data from a measurement, and a rotational correction for the data: | It is also possible to give a path for a file exported from 3D Explore with contour data from a measurement, and a rotational correction for the data: | ||
PoleFigureAngles([1 1 1; 0 0 1],[3 1 1; 1 3 3],'\Au_thin_inplanepole_WF1_m2.txt',30.9) | {|style="margin:auto;" | ||
|PoleFigureAngles([1 1 1; 0 0 1],[3 1 1; 1 3 3],'\Au_thin_inplanepole_WF1_m2.txt',30.9) | |||
|} | |||
Which will give the following table and pictures, without a file input only the table and the first graph will be given: | Which will give the following table and pictures, without a file input only the table and the first graph will be given: | ||
Revision as of 15:30, 29 November 2018
Feedback to this page: click here
XRD SmartLab
The Rigaku SmartLab is an advanced XRD for measuring on thin films. All thin films can be measured without fixating the sample, as the system has a so called In-Plane arm.
The user manual(s), user APV(s), technical information, and contact information can be found in LabManager:
Process information
XRR
With X-Ray Reflectivity measurements, it is possible to obtain information on thickness, density, and both surface and interface roughness on thin films. The technique does not depend on crystal structure and can be used on both amorphous, poly-, and single-crystalline materials. For film thickness measurement, films op to around 100 nm can be measured. You are welcome to try it on thicker films, but please confirm the measurement the first time by use of other equipment. XRR is a special case of a Theta/2Theta measurement. Rigaku gives a good explanation of the principles behind the XRR in this paper.
Theta-2Theta
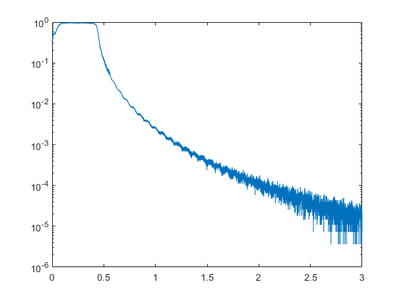
A theta-2theta scan can be used for identifying peaks and hence lattice constants and possible crystal orientations. It will also be possible to say something about the materials of the crystal, but it is not suitable for identification of composition. If a multilayers structure is, present a simulation of the stricter and comparison to the measurement can help identify layer thicknesses, atomic ratios, and interface roughness’s.
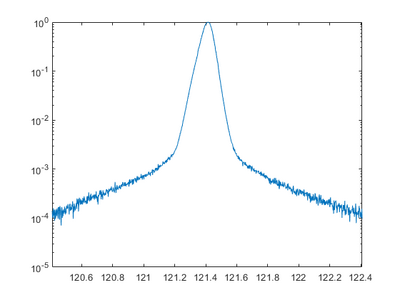
Rocking curve
A rocking curve is measured by fixing the 2Theta angle and changing the incident angle omega, which is the same as rocking the sample in the setup. This can be used to determine the preferred orientation and the degree of orientation of the measured material. For a perfect crystal and rocking curve will show a single sharp peak, if the layers are not perfect crystalline the diffraction peak will broaden. If an epitaxial layer is grown on a preface substrate, and the sample alignment is done to the substrate peak, a shift in omega will indicate a tilt in the crystal planes. A broadening could mean a dislocations, mosaicity, or curvature of the sample.
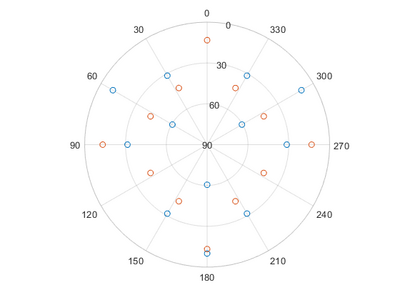
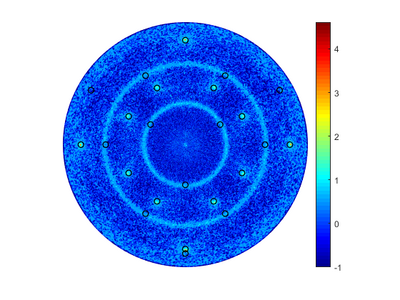
Pole figure
Pole figures can be used to determine the orientation of a crystal. For polycrystalline materials, it is possible to determine if there is a preferred direction of the crystal grains. In a pole figure, a measurement of a predefined peek for the material is measured in the half sphere above the sample. This will result in a map of intensities in relation to the angles from the surface normal and along sample rotation. For instance if you have a single crystalline sample with a [0 0 1] surface and measure on the [1 1 1] lattice plane, you should find 4 peaks spaced 90° in beta and at 35.26° in alpha.
Peak angles can be calculated by simple vector calculations; however, I have made a small MATLAB script calculating them for you. To use the script open MATLAB and call the program with two vectors as input. First vector should be your surface orientation, second input the plane you measure on. For a [1 1 1] substrate surface and a [3 1 1] plane and a [0 0 1] substrate surface and a [1 3 3] plane, where the measurement planes are dependent on the 2theta angle, the command looks like this:
| PoleFigureAngles([1 1 1; 0 0 1],[3 1 1; 1 3 3]) |
It is also possible to give a path for a file exported from 3D Explore with contour data from a measurement, and a rotational correction for the data:
| PoleFigureAngles([1 1 1; 0 0 1],[3 1 1; 1 3 3],'\Au_thin_inplanepole_WF1_m2.txt',30.9) |
Which will give the following table and pictures, without a file input only the table and the first graph will be given:
| Alpha | Beta | Alpha | Beta |
|---|---|---|---|
| 10.02 | 60.00 | 13.26 | 0.00 |
| 10.02 | 300.00 | 13.26 | 270.00 |
| 10.02 | 180.00 | 13.26 | 90.00 |
| 31.48 | 30.00 | 13.26 | 180.00 |
| 31.48 | 330.00 | 43.49 | 333.43 |
| 31.48 | 90.00 | 43.49 | 296.57 |
| 31.48 | 270.00 | 43.49 | 26.57 |
| 31.48 | 150.00 | 43.49 | 243.43 |
| 31.48 | 210.00 | 43.49 | 63.43 |
| 60.50 | 60.00 | 43.49 | 206.57 |
| 60.50 | 300.00 | 43.49 | 116.57 |
| 60.50 | 180.00 | 43.49 | 153.43 |
Rigaku describes Pole figures this paper.
Software for analysis
The software packages used for data analysis are available on the equipment computer, but we recommend that you install it on your personal computer. To run the software you need a USB dongle with a license on, these can be borrowed from Rebecca and Kristian in room 347-077. We only have 9 dongles available, so when you are done please return the dongle to danchip.
The software can be found on "CleanroomDrive\_Equipment\XRD\Rigaku software\RILauncher", it should be possible to install the software without a dongle. To use the software you have to log in. The user is: Administrator. There is no password.
| Equipment | XRD SmartLab | |
|---|---|---|
| Purpose | Crystal structure analysis and thin film thickness measurement |
|
| X-ray generator |
Maximum rated output |
3 kW |
|
Rated tube voltage |
20 to 45 kV | |
|
Rated tube current |
2 to 60 mA | |
|
Type |
Sealed tube | |
|
Target |
Cu | |
|
Focus size |
0.4x8 mm (Line/Point) | |
| Goniometer |
Scanning mode |
incident / receiver coupled or independent |
|
Goniomenter radius |
300 mm | |
|
Minimum step size |
0.0001° (0.36") | |
|
Sample stage |
| |
|
Sample size |
Diameter: 150 mm Thickness: 0~21 mm | |
| Optics | Incident side |
|
| Receiver side |
| |
| Substrates | Substrate size |
up to 150 mm wafers |
| Allowed materials |
All materials | |